SWGDAM Interpretation Guidelines
SWGDAM Interpretation Guidelines for Autosomal STR Typing by Forensic DNA Testing Laboratories
The Scientific Working Group on DNA Analysis Methods (SWGDAM) is a group of approximately 50 scientists representing federal, state, and local forensic DNA laboratories in the United States and Canada. During meetings, which are held twice a year, subcommittees discuss topics of interest to the forensic DNA community and often develop documents to provide direction and guidance for the community. A mixture interpretation subcommittee was formed in January 2007 and worked for several years to provide a guidance document on autosomal short tandem repeat (STR). This document was presented to the full SWGDAM group and received approval in January 2010.
The document provides guidelines for the interpretation of DNA typing results from short tandem repeats (STR) and supersedes the Scientific Working Group on DNA Analysis Methods (SWGDAM) Short Tandem Repeat (STR) Interpretation Guidelines (2000). The revised guidelines are not intended to be applied retroactively. Guidance is provided for forensic casework analyses on the identification and application of thresholds for allele detection and interpretation, and appropriate statistical approaches to the interpretation of autosomal STRs with further guidance on mixture interpretation. Laboratories are encouraged to review their standard operating procedures and validation data in light of these guidelines and to update their procedures as needed. It is anticipated that these guidelines will evolve further as future technologies emerge. Some aspects of these guidelines may be applicable to low level DNA samples. However, this document is not intended to address the interpretation of analytical results from enhanced low template DNA techniques.
Introduction
The interpretation of DNA typing results for human identification purposes requires professional judgment and expertise. Additionally, laboratories that analyze DNA samples for forensic casework purposes are required by the Quality Assurance Standards for Forensic DNA Testing Laboratories (effective September 1, 2011) to establish and follow documented procedures for the interpretation of DNA typing results and reporting. Due to the multiplicity of forensic sample types and the potential complexity of DNA typing results, it is impractical and infeasible to cover every aspect of DNA interpretation by a preset rule. However, the laboratory should utilize written procedures for interpretation of analytical results with the understanding that specificity in the standard operating protocols will enable greater consistency and accuracy among analysts within a laboratory. It is recommended that standard operating procedures for the interpretation of DNA typing results be sufficiently detailed that other forensic DNA analysts can review, understand in full, and assess the laboratory’s policies and practices. The laboratory’s interpretation guidelines should be based upon validation studies, scientific literature, and experience.
Background
Upon completion of the technical aspects of DNA analysis, DNA typing results must be verified and interpreted. The verification of the accuracy of the DNA typing results involves a review of peak designations and other software-generated information, as well as an evaluation of quality controls. Based on this assessment, the DNA analyst performs interpretations, makes comparisons among samples (where appropriate), and draws conclusions. These data and conclusions are technically reviewed, and the conclusions are typically captured for documentation and communication purposes within a laboratory report.
Using current technologies for human identification, DNA typing results are derived through application of analytical software during and after electrophoresis of fluorescently-labeled amplification products that are generated for each sample using an amplification kit. For each sample, the software translates fluorescence intensity data into electropherograms and then labels any detected peaks with such descriptors as size (in base-pairs, or bp) and peak height (in relative fluorescence units, or RFU). Using allelic ladders for reference, the software then labels peaks that meet certain criteria with allelic designations.
To ensure the accuracy of these computer-generated allele designations, the DNA analyst must verify that appropriate genotyping parameters (i.e., internal size standard and allelic ladder) were used and that the correct genotyping results were obtained for a known positive control. Additionally, if a sample is amplified using multiple kits that contain redundant loci, the DNA analyst must address the concordance of the genotyping results at the loci that are common to both kits. As an example, a given sample amplified using both the Profiler PlusTM and COfilerTM Amplification Kits exhibits concordance when identical alleles for the genetic loci amelogenin, D3S1358, and D7S820 are obtained. After verification of the allelic designations, the alleles are classified based on their peak height relative to an established minimum peak height threshold for comparison purposes.
The results of the analysis controls—i.e., reagent blank(s), positive amplification control(s), and negative amplification control(s)—are evaluated. If the reagent blank(s), positive amplification control(s), and negative amplification control(s) yield results that are within their prescribed specifications, the DNA analyst interprets the DNA typing results from each sample to determine if the DNA typing results originated from a single donor or multiple donors. If the expected results are not obtained from a control sample(s), the DNA analyst must determine if the control(s) and/or sample(s) should be re-processed or proceed within the prescribed limitations of interpretation.
Based on the interpretation of the forensic samples and a comparison of the DNA typing results obtained from the questioned sample(s) to those of any known sample(s), or a comparison between multiple questioned samples, a DNA analyst can reach one of three primary conclusions: cannot exclude, can exclude, or inconclusive/uninterpretable.
Statistical interpretation for reported inclusionary results provides weight to the inclusionary statement. Statistical analysis is not required for exclusionary conclusions, comparisons between multiple questioned samples without a comparison to a known sample, nor applicable to inconclusive/uninterpretable results. The conclusions reached as part of the DNA interpretation process are compiled into a written draft by the DNA analyst and are subjected to technical and administrative reviews prior to issuing a final case report.
This document addresses definitions, data evaluation, interpretation of results, and conclusions/reporting for autosomal STR typing, including guidance on mixture interpretation. Approaches to statistical interpretation are presented. A list of relevant literature is also included to provide further source material.
1. Preliminary Evaluation of Data
The laboratory should develop criteria to determine whether an instrumental response represents the detection of DNA fragment(s) rather than instrument noise. An analytical threshold defines the minimum height requirement at and above which detected peaks can be reliably distinguished from background noise. Because the analytical threshold is based upon a distribution of noise values, it is expected that occasional, non-reproducible noise peaks may be detected above the analytical threshold. An analytical threshold should be sufficiently high to filter out noise peaks. Usage of an exceedingly high analytical threshold increases the risk of allelic data loss which is of potential exclusionary value.
1.1 Analytical threshold:
The Laboratory should establish an analytical threshold based on signal-to-noise analyses of internally derived empirical data. As an example, an analytical threshold may be based on two times the intensity difference between the highest peak and lowest trough within the instrumental noise data. Other scientific methods may be used. The usage of an analytical threshold value that differs substantially from manufacturer’s recommendations should be supported by internal signal-to-noise assessments.
1.2 The laboratory must develop criteria to evaluate internal standards and/or allelic ladders.
1.3 Controls are required to assess analytical procedures.
1.3.1 The laboratory must establish criteria for evaluation of the following controls, including but not limited to: reagent blank and positive and negative amplification controls.
1.3.2 The laboratory must develop criteria for the interpretation and documentation of results in the event that the controls do not perform as expected.
1.4 A laboratory using STR multiplexes that contain redundant loci must establish criteria regarding the concordance of such data.
2. Allele Designation
2.1 The laboratory establishes criteria to assign allele designations to appropriate peaks.
2.1.1 Locus Designation: The laboratory establishes criteria to address locus assignment for alleles. The criteria should address alleles that fall above the largest or below the smallest allele (or virtual bin) of the allelic ladder.
2.1.2 Allele Designation: The laboratory designates alleles as numerical values in accordance with recommendations of the International Society of Forensic Genetics.
2.1.2.1 Allele designation is based operationally on the number of repeat sequences contained within the allele and by comparison to an allelic ladder.
2.1.2.2 The laboratory establishes guidelines for the designation of alleles containing an incomplete repeat motif (i.e., an off-ladder allele falling within the range spanned by the ladder alleles). This designation includes the number of complete repeats and, separated by a decimal point, the number of base pairs in the incomplete repeat (e.g., FGA 18.2 allele).
2.1.2.3 The laboratory establishes criteria for designating alleles that fall above the largest or below the smallest allele of the allelic ladder (or virtual bin). Extrapolation of an above/below ladder allele to a specific designation (e.g., generally to no more than one repeat unit) should also be supported by precision studies, validation, and determination of measurement variance. Above/below ladder alleles should be designated as either greater than (>) or less than (<) the respective ladder allele (or virtual bin), or designated numerically when appropriate extrapolation can be used. When the “>” or “<” designation is used, the laboratory should establish criteria, based on relative sizes, for the comparison of such alleles among samples.
3. Interpretation of DNA Typing Results
3.1 Non-Allelic Peaks:
Because forensic DNA typing characterizes STR loci using PCR and electrophoretic technologies, some data that result from this analytical scheme may not represent actual alleles that originate in the sample. It is therefore necessary, before the STR typing results can be used for comparison purposes, to identify any potential non-allelic peaks. Non-allelic peaks may be PCR products (e.g., stutter, non-template dependent nucleotide addition, and non-specific amplification product), analytical artifacts (e.g., spikes and raised baseline), instrumental limitations (e.g., incomplete spectral separation resulting in pull-up or bleed-through), or may be introduced into the process (e.g., disassociated primer dye). Generally, non-allelic data such as stutter, non-template dependent nucleotide addition, disassociated dye, and incomplete spectral separation are reproducible; spikes and raised baseline are generally non-reproducible.
3.1.1 The laboratory establishes criteria based on empirical data (obtained internally or externally), and specific to the amplification and detection systems used, to address the interpretation of non-allelic peaks. The guidelines address identification of non-allelic peaks and the uniform application, across all loci of a DNA profile, of the criteria used to identify non-allelic peaks.
3.1.1.1 In general, the empirical criteria are based on qualitative and/or quantitative characteristics of peaks. As an example, dye artifacts and spikes may be distinguished from allelic peaks based on morphology and/or reproducibility. Stutter and non-template dependent nucleotide addition peaks may be characterized based on size relative to an allelic peak and amplitude.
3.1.1.2 While the application of an analytical threshold may serve to filter out some non-allelic peaks, the analytical threshold should be established based on signal-to-noise considerations (i.e., distinguishing potential allelic peaks from background). The analytical threshold should not be established for purposes of avoiding artifact labeling, as such may result in the potential loss of allelic data.
3.1.1.3 The laboratory establishes guidelines addressing off-scale data. Fluorescence detection instruments have a limited linear range of detection, and signal saturation can result in off-scale peaks. Following peak detection, such peaks in the analyzed data are assigned an artificial height value which is not representative of the true amplitude. Peak height values for off-scale peaks should not be used in quantitative aspects of interpretation (e.g., stutter and peak height ratio assessments).
3.2 Application of Peak Height Thresholds to Allelic Peaks:
Amplification of low-level DNA samples may be subject to stochastic effects, where two alleles at a heterozygous locus exhibit considerably different peak heights (i.e., peak height ratio generally <60%) or an allele fails to amplify to a detectable level (i.e., allelic dropout). Stochastic effects within an amplification may affect one or more loci irrespective of allele size. Such low-level samples exhibit peak heights within a given range which is dependent on quantitation system, amplification kit, and detection instrumentation. A threshold value can be applied to alert the DNA analyst that all of the DNA typing information may not have been detected for a given sample. This threshold, referred to as a stochastic threshold, is defined as the value above which it is reasonable to assume that allelic dropout has not occurred within a single-source sample. The application of a stochastic threshold to the interpretation of mixtures should take into account the additive effects of potential allele sharing.
3.2.1 The laboratory establishes a stochastic threshold based on empirical data derived within the laboratory and specific to the quantitation and amplification systems (e.g., kits) and the detection instrumentation used. It is noted that a stochastic threshold may be established by assessing peak height ratios across multiple loci in dilution series of DNA amplified in replicate. The RFU value above which it is reasonable to assume that, at a given locus, allelic dropout of a sister allele has not occurred constitutes a stochastic threshold.
3.2.1.1 If measures are used to enhance detection sensitivity (i.e., allelic height), the laboratory should perform additional studies to establish independent criteria for application of a separate stochastic threshold(s). Such measures may include, but not be limited to, increased amplification cycle number, increased injection time, and post-amplification purification/concentration of amplified products.
3.2.1.2 For samples for which an assumption can be made as to the number of contributors, the laboratory should establish criteria for comparison of allelic peaks which fall below the stochastic threshold. As an example, if a locus in an assumed single-source sample exhibits two peaks, one or both of which are below the stochastic threshold, the laboratory may use that locus for comparison purposes. Also, the presence of male DNA may be established based on a Y-allele at amelogenin that is below the stochastic threshold.
3.2.2 If a stochastic threshold based on peak height is not used in the evaluation of DNA typing results, the laboratory must establish alternative criteria (e.g., quantitation values or use of a probabilistic genotype approach) for addressing potential stochastic amplification. The criteria must be supported by empirical data and internal validation and must be documented in the standard operating procedures.
3.3 Peak Height Ratio:
Intra-locus peak height ratios (PHR) are calculated for a given locus by dividing the peak height of an allele with a lower RFU value by the peak height of an allele with a higher RFU value, and then multiplying this value by 100 to express the PHR as a percentage.
3.3.1 The laboratory should establish PHR requirements based on empirical data for interpretation of DNA typing results from single-source samples. Different PHR expectations can be applied to individual loci (e.g., 70% for D3S1358, 65% for vWA, etc.); alternatively, a single PHR expectation can be applied to multiple loci (e.g., 60%).
3.3.1.1 The laboratory may evaluate PHRs at various DNA template levels (e.g., dilution series of DNA). It is noted that different PHR expectations at different peak height ranges may be established.
3.3.2 PHR requirements are only applicable to allelic peaks that meet or exceed the stochastic threshold.
3.4 Number of Contributors to a DNA Profile:
Generally, a sample is considered to have originated from a single individual if one or two alleles are present at all loci for which typing results were obtained (although tri-allelic loci may occur), and the peak height ratios for all heterozygous loci are within the empirically determined values. It is noted that peak height imbalances may be seen in the typing results from, for example, a primer binding site variant that results in attenuated amplification of one allele of a heterozygous pair.
A sample is generally considered to have originated from more than one individual if three or more alleles are present at one or more loci (excepting tri-allelic loci) and/or the peak height ratios between a single pair of allelic peaks for one or more loci are below the empirically determined heterozygous peak height ratio expectation. Generally, the minimum number of contributors to a mixed sample can be determined based on the locus that exhibits the greatest number of allelic peaks. As an example, if at most five alleles are detected per locus, then the DNA typing results are consistent with having arisen from at least three individuals.
3.4.1 For DNA mixtures, the laboratory should establish guidelines for determination of the minimum number of contributors to a sample. Alleles need not meet the stochastic threshold to be used in this assessment.
3.4.2 The laboratory should define the number of alleles per locus and the relative intra-locus peak height requirements for assessing whether a DNA typing result is consistent with originating from one or more sources. The minimum number of loci should be defined for determination of whether a sample is a mixture.
3.4.3 Where multiple amplifications and/or injections are generated for a given sample extract, the laboratory should establish guidelines for determining which results are used for comparisons and statistical calculations.
3.4.3.1 If composite profiles (i.e., generated by combining typing results obtained from multiple amplifications and/or injections) are used, the laboratory should establish guidelines for the generation of the composite result. When separate extracts from different locations on a given evidentiary item are combined prior to amplification, the resultant DNA profile is not considered a composite profile. Unless there is a reasonable expectation of sample(s) originating from a common source (e.g., duplicate vaginal swabs or a bone), allelic data from separate extractions from different locations on a given evidentiary item should not be combined into a composite profile. The laboratory should establish guidelines for determining the suitability of developing composite profiles from such samples.
3.5 Interpretation of DNA Typing Results for Mixed Samples:
An individual’s contribution to a mixed biological sample is generally proportional to their quantitative representation within the DNA typing results. Accordingly, depending on the relative contribution of the various contributors to a mixture, the DNA typing results may potentially be further refined.
As an example, if a sample contains a predominance of one individual’s DNA, that individual’s DNA profile may be determined. This state results in a distinguishable mixture, whereby there is a distinct contrast in signal intensities (e.g., peak heights) among the different contributors’ alleles. In such instances, major and/or minor contributors may be determined. Discernment of the STR typing results for the major or minor contributors to a mixture may be limited to only some loci (with the remaining loci yielding multiple potential genotypes for the major or minor contributor). The major (and possibly the minor) contributor may effectively constitute a deduced single-source profile.
Alternatively, if the amounts of biological material from multiple donors are similar, it may not be possible to further refine the mixture profile. When major or minor contributors cannot be distinguished because of similarity in signal intensities, the sample is considered to be an indistinguishable mixture. The classification as indistinguishable may be limited to some, not all, of the loci for which DNA typing results are obtained and does not imply that the profile is uninterpretable. Individuals may still be included or excluded as possible contributors to an indistinguishable mixture.
Evidence items taken directly from an intimate sample, as determined by the laboratory, are generally expected to yield DNA from the individual from whom the sample was taken. If another source of DNA is present in sufficient quantity in such a sample, a mixture of DNA is likely to be detected. Based on this expectation, any DNA typing results from such a mixture that match a conditional known sample (e.g., from the victim) may be separated from the other mixture results to facilitate identification of the foreign alleles. The obligate alleles may effectively constitute a single-source profile (i.e., if there is one DNA contributor in addition to the individual from whom the sample was taken) or a mixture profile (i.e., if there are multiple additional DNA contributors). A similar state can exist when another known individual (i.e., consensual partner) is expected to have contributed biological material to the mixed sample.
3.5.1 The laboratory should establish guidelines based on peak height ratio assessments for evaluating potential sharing of allelic peaks among contributors and for determining whether contributors to a mixed DNA typing result are distinguishable. When assessing peak height ratios, pair-wise comparison of all potential genotypic combinations should be evaluated.
3.5.2 The laboratory should define and document what, if any, assumptions are used in a particular mixture deconvolution.
3.5.2.1 If no assumptions are made as to the number of contributors, at a minimum, the laboratory should assign to a major contributor an allele (e.g., homozygous) or pair of alleles (e.g., heterozygous) of greater amplitude at a given locus that do not meet peak height ratio expectations with any other allelic peak(s).
3.5.2.2 If assumptions are made as to the number of contributors, additional information such as the number of alleles at a given locus and the relative peak heights can be used to distinguish major and minor contributors.
3.5.3 A laboratory may define other quantitative characteristics of mixtures (e.g., mixture ratios) to aid in further refining the contributors.
3.5.3.1 Differential degradation of the contributors to a mixture may impact the mixture ratio across the entire profile.
3.5.4 Mixtures with a Single Major Contributor and One or More Minor Contributors:
3.5.4.1 In general, heterozygous alleles attributed to a major contributor should meet the laboratory’s established peak height ratio expectations for single-source samples. Due to the potential for overlapping peaks to cause imbalance of major heterozygous alleles, the laboratory may establish a quantitative means of evaluating the distinction in peak heights of the major and minor contributors (i.e., mixture ratio).
3.5.4.2 After deconvolution, the DNA typing results attributed to an individual minor contributor should also meet PHR expectations. The PHR expectations of a minor contributor may be reduced due to stochastic peak height variation and the additive effects of peak sharing (e.g., minor peak and stutter peaks).
3.5.4.3 Due to the possibility that the minor contributor’s alleles may be shared by the major contributor (and thus masked), determination of a single genotype for a minor contributor may be possible at only some loci (while multiple allelic combinations, or allelic drop out, are possible at other loci).
3.5.5 Mixtures with Multiple Major Contributors and One or More Minor Contributors: The laboratory should establish guidelines based on peak height ratio assessments and/or mixture ratios for determining whether multiple major contributors are present in a mixed sample.
3.5.6 Mixtures with Indistinguishable Contributors: The laboratory should establish guidelines based on peak height ratio assessments for identifying mixtures for which no major or minor contributors can be discerned.
3.5.7 Mixtures with a Known Contributor(s): The laboratory should establish guidelines for determining whether separation of a known contributor’s profile is applicable (e.g., based on the types of evidentiary items).
3.5.7.1 At a minimum, where there is no indication of sharing of the known and obligate alleles, the laboratory should separate out those alleles attributable to the known sample (e.g., victim, consensual partner, etc.).
3.5.7.2 To further refine the obligate alleles in a profile, the laboratory may establish guidelines for addressing potential sharing of alleles among the individual known to have contributed to a sample and the additional contributor(s).
3.5.8 Interpretation of Potential Stutter Peaks in a Mixed Sample:
3.5.8.1 For mixtures in which minor contributors are determined to be present, a peak in stutter position (generally n-4) may be determined to be 1) a stutter peak, 2) an allelic peak, or 3) indistinguishable as being either an allelic or stutter peak. This determination is based principally on the height of the peak in the stutter position and its relationship to the stutter percentage expectations established by the laboratory.
3.5.8.2 Generally, when the height of a peak in the stutter position exceeds the laboratory’s stutter expectation for a given locus, that peak is consistent with being of allelic origin and should be designated as an allele.
3.5.8.3 If a peak is at or below this expectation, it is generally designated as a stutter peak. However, it should also be considered as a possible allelic peak, particularly if the peak height of the potential stutter peak(s) is consistent with (or greater than) the heights observed for any allelic peaks that are conclusively attributed (i.e., peaks in non-stutter positions) to the minor contributor(s).
3.6 Comparison of DNA Typing Results:
The following determinations can be made upon comparison of evidentiary and known DNA typing results (and between evidentiary samples):
- The known individual cannot be excluded (i.e., is included) as a possible contributor to the
DNA obtained from an evidentiary item. - The known individual is excluded as a possible contributor.
- The DNA typing results are inconclusive/uninterpretable.
- The DNA typing results from multiple evidentiary items are consistent or inconsistent with originating from a common source(s).
3.6.1 The laboratory must establish guidelines to ensure that, to the extent possible, DNA typing results from evidentiary samples are interpreted before comparison with any known samples, other than those of assumed contributors.
3.6.2 DNA typing results may not be obtained at all loci for a given evidentiary sample (e.g., due to DNA degradation, inhibition of amplification, and/or low-template quantity); a partial profile thus results.
3.6.2.1 For partial profiles, the determination of which alleles/loci are suitable for comparison and statistical analysis should be made prior to comparison to the known profiles.
3.6.2.2 The laboratory should establish guidelines for inclusions and exclusions when a known individual’s DNA profile is not fully observed in the evidentiary profile.
3.6.3 The laboratory must establish guidelines for inclusionary, exclusionary, and inconclusive/uninterpretable conclusions based on comparisons of DNA typing results from known samples and both single-source and mixed evidentiary samples.
3.6.4 For mixtures for which two or more individuals cannot be excluded as potential contributors, the laboratory may establish guidelines for assessing whether all of the DNA typing results obtained from the mixed sample are accounted for by the multiple known samples.
3.6.5 Because assumptions regarding the origin of evidence or the number of contributors to a mixture can impact comparisons, the laboratory should establish guidelines for documenting any assumptions that are made when formulating conclusions.
3.6.6 The laboratory should establish guidelines for identifying DNA typing results for which comparisons of evidentiary and known samples are not made (at a minimum, to include inconclusive/uninterpretable results).
4. Statistical Analysis of DNA Typing Results
In forensic DNA testing, calculations are performed on evidentiary DNA profiles that are established as relevant in the context of the case to aid in the assessment of the significance of an inclusion. These calculations are based on the random match probability (RMP), the likelihood ratio (LR), or the combined probability of exclusion/inclusion (CPE/CPI).
While the RMP is commonly thought of in terms of single-source profiles, the application of this formula to evidentiary profiles inherently includes an assumption of the number of contributors to the DNA sample. As such, this document also applies the term RMP to mixture calculations where the number of contributors is assumed (this has sometimes been referred to as a “modified RMP”). By using the RMP nomenclature, these calculations are distinguished from the CPI nomenclature which is commonly thought of in terms of a mixture calculation that makes no assumption as to the number of contributors.
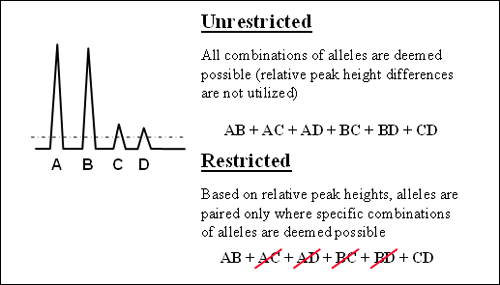
In addition to assumptions of the number of contributors, quantitative peak height information and mixture ratio assessments may or may not be included in the interpretation of an evidentiary profile. Calculations performed using interpretations incorporating this information are termed “restricted.” When this quantitative peak height information is not included, the resultant calculation is termed “unrestricted” (Figure 1).
Figure 1. Illustration of “restricted” versus “unrestricted” approaches based on relative peak heights (using an assumption of two donors with all peaks above the stochastic threshold).
The genetic loci and assumptions used for statistical calculations must be documented, at a minimum, in the case notes.
4.1 The laboratory must perform statistical analysis in support of any inclusion that is determined to be relevant in the context of a case, irrespective of the number of alleles detected and the quantitative value of the statistical analysis.
4.1.1 The laboratory should establish guidelines where multiple stains from the same or separate items have provided genetic information that is consistent with originating from a common source(s) but having various levels of discrimination. In general, the statistics for the typing results that provide the most genetic information and/or the highest discrimination potential are reported.
4.2 For calculating the CPE or RMP, any DNA typing results used for statistical analysis must be derived from evidentiary items and not known samples. This precludes combining multiple CPE or RMP results for the same mixture component of an evidentiary sample. However, different calculations may be made for the same mixture component if different assumptions as to the number of contributors are made and clearly stated in the case notes and/or report.
4.3 The laboratory must not use inconclusive/uninterpretable data (e.g., at individual loci or an entire multi-locus profile) in statistical analysis.
4.3.1 For a distinguishable mixture, a major contributor(s) profile may be suitable for statistical analysis even in the presence of inconclusive minor contributor results.
4.4 Exclusionary conclusions do not require statistical analysis.
4.5 The laboratory must document the source of the population database(s) used in any statistical analysis.
4.6 The formulae used in any statistical analysis must be documented and must address both homozygous and heterozygous typing results, multiple locus profiles, mixtures, minimum allele frequencies, and, where appropriate, biological relationships.
4.6.1 Given a profile for which multiple formulae are applicable, the laboratory must have guidelines for the selection of the formula(e) suitable for statistical application (see Table 1).
4.6.2 It is not appropriate to calculate a composite statistic using multiple formulae for a multi-locus profile. For example, the CPI and RMP cannot be multiplied across loci in the statistical analysis of an individual DNA profile because they rely upon different fundamental assumptions about the number of contributors to the mixture.
4.6.3 When using CPE/CPI (with no assumptions of number of contributors) to calculate the probability that a randomly selected person would be excluded/included as a contributor to the mixture, loci with alleles below the stochastic threshold may not be used for statistical purposes to support an inclusion. In these instances, the potential for allelic dropout raises the possibility of contributors having genotypes not encompassed by the interpreted alleles.
4.6.3.1 Alleles below the stochastic threshold may be used for comparisons and/or to establish the presence of a mixture or male DNA (e.g., Y allele at amelogenin).
4.6.3.2 A restricted CPE/CPI may be applied to multiple major contributors despite the presence of minor contributor(s) alleles below the stochastic threshold; a description of how to calculate can be found in Section 5.3.5.
4.7 If a laboratory uses source attribution statements, then it must establish guidelines for the criteria on which such a declaration is based.
5. Statistical Formulae
5.1 Whenever the statistical analysis at a locus is meant to represent all possible contributors to a mixture, if there is a reasonable possibility that locus dropout could have led to the loss of an entire genotype, then a statistical calculation should not be performed for that locus. Similarly, the product rule should not be applied when the resultant set of combined profiles would not include all individuals who would not be excluded as possible contributors to the mixture.
5.2 Random Match Probability (RMP):
5.2.1 When the interpretation is based upon the assumption of a single contributor (or a single major contributor to a mixture), the RMP formulae are those described in NRCII recommendations 4.1, 4.2, 4.3, and 4.4. The most commonly used formulae are listed below:
5.2.1.1 For heterozygote genotypes, the formula is 2pq. This is NRCII formula 4.1b.
5.2.1.2 For homozygote genotypes, the formula is p2 + p(1-p)θ, where q = 0.01 or 0.03 in accordance with NRCII. This is NRCII formula 4.4a.
5.2.1.3 For single-allele profiles where the zygosity is in question (e.g., it falls below the stochastic threshold):
5.2.1.3.1 The formula 2p, as described in recommendation 4.1 of NRCII, may be applied to this result.
5.2.1.3.2 Instead of using 2p, the algebraically identical formulae 2p – p2 and p2 + 2p(1-p) may be used to address this situation without double-counting the proportion of homozygotes in the population.
5.2.1.3.3 Laboratories may choose to assign the value of 1 to the scenario described in 5.2.1.3., i.e. not use the locus for statistical weight.
5.2.1.4 Conditional subpopulation calculations may also be performed in accordance with NRCII formulae 4.10a and 4.10b.
5.2.2 When the interpretation is conditioned upon the assumption of a particular number of contributors greater than one, the RMP is the sum of the individual frequencies for the genotypes included following a mixture deconvolution. Examples are provided below:
5.2.2.1 In a sperm fraction mixture (at a locus having alleles P, Q, and R) assumed to be from two contributors, one of whom is the victim (having genotype QR), the sperm contributor genotypes included post-deconvolution might be PP, PQ, and PR. In this case, the RMP for the sperm DNA contributor could be calculated as [p2 + p(1-p)θ] + 2pq + 2pr.
5.2.2.2 In a sperm fraction mixture (at a locus having alleles P, Q, and R) assumed to be from two contributors, where the major contributor is the victim (having genotype QR), there remains an obligate minor contributor P allele above the stochastic threshold. Also present in the results are two peaks filtered as possible stutter (S* and T*). If both filtered peaks are within an RFU range that could reasonably be paired with the P allele as heterozygous genotypes, the sperm contributor genotypes included post-deconvolution might be PP, PQ, PR, PS*, and PT*. In this case, the RMP for the sperm DNA contributor could be calculated as [p2 + p(1-p)θ] + 2pq + 2pr + 2ps + 2pt. Some laboratories might instead choose to apply a single-allele formula as discussed in section 5.2.1.3, e.g., 2p.
5.2.2.3. In a mixture having at a locus alleles P, Q, and R, assumed to be from two contributors, where all three alleles are below the stochastic threshold, the interpretation may be that the two contributors could be a heterozygote-homozygote pairing where all alleles were detected, a heterozygote-heterozygote pairing where all alleles were detected, or a heterozygote-heterozygote pairing where a fourth allele might have dropped out. In this case, the RMP must account for all heterozygotes and homozygotes represented by these three alleles, but also all heterozygotes that include one of the detected alleles. The RMP for this interpretation could be calculated as
(2p – p2) + (2q – q2) + (2r – r2) – 2pq – 2pr – 2qr.
5.2.2.3.1 Since 2p includes 2pq and 2pr, 2q includes 2pq and 2qr, and 2r includes 2pr and 2rq, the formula in 5.2.2.3 subtracts 2pq, 2pr, and 2qr to avoid double-counting these genotype frequencies.
5.2.2.3.2 Laboratories may choose to use the formula 2p + 2q + 2r for the scenario described in 5.2.2.3.
5.2.2.3.3 Laboratories may choose to assign the value of 1 to the scenario described in 5.2.2.3, i.e. not use the locus for statistical weight.
5.2.2.4 Care should be taken to not report a calculated RMP greater than 1.0. This can occur when using the calculations discussed in 5.2.2.1 and 5.2.2.2 (due to the application of q in the standard homozygote formula but not in the heterozygote formula) and in 5.2.2.3.1 (due to the double counting of the PP, QQ, RR, PQ, PR, and QR genotype frequencies).
5.2.2.5 In a sperm fraction assumed to be from two contributors, one of whom is the victim, the sperm contributor genotypes included post-deconvolution might include only a single genotype (PQ) at locus 1, but multiple possible genotypes (UU or UV) at locus 2. In this case, the two-locus RMP for the sperm DNA contributor could be calculated as 2pq * [u2 + u(1-u)θ + 2uv].
5.2.2.6 The unrestricted RMP might be calculated for mixtures that display no indications of allelic dropout. The formulae include an assumption of the number of contributors, but relative peak height information is not utilized. For two-person mixtures, the formulae for loci displaying one, two, or three alleles are identical to the CPI calculation discussed in section 5.3. For loci displaying four alleles (P, Q, R, and S), homozygous genotypes would not typically be included. The unrestricted RMP in this case would require the subtraction for homozygote genotype frequencies, e.g., (p + q + r + s)2 – p2 – q2 – r2 – s2.
5.2.3 When a suspect’s profile has been determined to match the unknown profile, if the alternate hypothesis is that a relative of the suspect is in fact the source of the unknown profile, then all efforts should be undertaken to obtain a sample directly from the relative in question so that there is no need to rely on a probability-based estimate of a coincidental match.
In the absence of a direct comparison, conditional match probabilities for various relatives can be calculated in accordance with NRCII formulae 4.8 and 4.9.
5.2.3.1 Full Siblings (NRCII formulae 4.9a and 4.9b)
|
Genotype of suspect
|
Probability of the same
genotype in a sibling |
|---|---|
|
PP
|
(1 + 2p + p2) / 4
|
|
PQ
|
(1 + p + q + 2pq) / 4
|
5.2.3.2 Other Relatives (NRCII formulae 4.8a and 4.8b)
|
Genotype of suspect
|
Probability of the same
genotype in a relative |
|---|---|
|
PP
|
p2 + 4p(1 – p)F
|
|
PQ
|
2pq + 2(p + q – 4pq)F
|
where F = 1/4 for parent and offspring; 1/8 for half-siblings; 1/8 for uncle and nephew; 1/8 for grandparent and grandchild; and 1/16 for first cousins.
5.2.3.3 Conditional subpopulation corrections could also be applied to these formulae following the methods of Ayres (2000) as described in Fung and Hu (2008).
5.3 Combined Probability of Inclusion (CPI) and Exclusion (CPE):
5.3.1 PI is calculated as (sum of allele frequencies)2 for each locus.
5.3.2 The CPI is the product of the individual locus PIs: CPI = PI1 * PI2 * ... * PIN
5.3.3 The PE has been commonly presented two ways:
5.3.3.1 PE = 1 – PI
5.3.3.2 PE = q2 + 2pq, where p is the sum of allele frequencies and q represents all other alleles (1 – p). This is analogous to the single allele formula described in 5.2.1.3.2.
5.3.3.3 Population substructure corrections can also be applied using PE = 1 – [p2 – p(1 – p)θ], where p is the sum of allele frequencies observed at that locus.
5.3.4 The CPE has been commonly presented two ways:
5.3.4.1 CPE = 1 – CPI
5.3.4.2 CPE = 1 – [(1 – PE1) * [(1 – PE2) * ... * (1 – PEN)]
5.3.5 The CPI and CPE are typically applied to all alleles detected in a mixture, subject to the limitations described in section 4.6.3. This section also allowed for a restricted CPI and CPE. Examples of both scenarios are provided below:
5.3.5.1 Unrestricted CPI and CPE. In a mixture at a locus having alleles P, Q, and R, all above the laboratory’s stochastic threshold, the interpretation might be that all potential contributors to this mixture have genotypes consisting of some combination of the detected alleles (PP, QQ, RR, PQ, PR, and QR). In this case, the probability of inclusion for the mixture could be calculated as (p + q + r)2.
5.3.5.2 Unrestricted CPI and CPE. In a mixture at a locus having alleles P, Q, R, and S where alleles P, Q, and R are above the stochastic threshold, but allele S is below that threshold, in the standard application of the CPI and CPE, no calculation would be performed at this locus.
5.3.5.3 Restricted CPI and CPE. Given (a) a mixture at a locus having alleles P, Q, R, and S, (b) alleles P, Q, and R significantly (as defined by the laboratory) above the stochastic threshold, and (c) allele S is below the stochastic threshold, the interpretation might be that the higher RFU alleles are a distinct group, separate from the contributor(s) of the low-RFU S allele. The lab might choose to calculate a restricted probability of inclusion utilizing just the P, Q, and R alleles, (p + q + r)2.
5.3.5.3.1 Based on the above example, had the S allele been greater than the stochastic threshold, but still identified as distinct from the higher-RFU alleles, a second general CPI or CPE could have been calculated using all four alleles.
5.4 Likelihood Ratio (LR):
5.4.1 When the evidence profile is determined to be single source, and the reference and evidence profiles are identical at all loci, LR = 1/RMP.
5.4.1.1 The numerator of the LR calculation would assume the suspect’s contribution, meaning that the probability of observing results consistent with his or her profile would be 1.0.
5.4.1.2 The denominator would assume that the suspect is not the contributor. The probability of a randomly selected person having the evidence profile is represented by the RMP.
5.4.2 The calculation of the LR in a mixture is dependent upon the evidence profile, the comparison reference profile(s), and the individual hypotheses. Given the myriad possible combinations, any list would be necessarily incomplete. A limited set of examples is provided below.
5.4.2.1 An “unrestricted” LR is the LR calculated without taking peak heights into consideration, especially in the denominator.
5.4.2.1.1 At a locus, a mixture with alleles P and Q, is assumed to be from two contributors, and displays no indications of allelic dropout. No further considerations of peak heights are undertaken. The suspect in question is PP, and no other reference standards are being considered for inclusion.
The numerator of the LR calculation would assume the suspect’s contribution, meaning that the probability of observing results consistent with his or her genotype would be 1.0. The second, unknown contributor must complete the mixture by having allele Q and nothing other than P or Q. Therefore the numerator to the calculation would be the sum of the frequencies for the second contributor’s possible genotypes (QQ and PQ).
LR numerator = [q2 + q(1-q)θ] + 2pq
The denominator of the LR calculation might assume that the mixture is a combination of two unknown contributors. Alternate hypotheses are possible as long as the numerator and denominator hypotheses are mutually exclusive. The unknown contributors must have no alleles other than P or Q, and the combination of their genotypes must complete the detected mixture of P and Q.
| Contrib. #1 | Contrib. #2 | Combined Probability |
|---|---|---|
|
PP
|
QQ
|
[p2 + p(1-p)θ] * [q2 + q(1-q)θ]
|
|
QQ
|
PP
|
[q2 + q(1-q)θ] * [p2 + p(1-p)θ]
|
|
PQ
|
PP
|
2pq * [p2 + p(1-p)θ]
|
|
PP
|
PQ
|
[p2 + p(1-p)θ] * 2pq
|
|
PQ
|
QQ
|
2pq * [q2 + q(1-q)θ]
|
|
QQ
|
PQ
|
[q2 + q(1-q)θ] * 2pq
|
|
PQ
|
PQ
|
2pq * 2pq
|
LR denominator = the sum of the possible combinations of genotypes (i.e., summing the seven combined probabilities).
5.4.2.2 A “restricted” LR is the LR calculated once relative peak heights are taken into consideration. Note: Within an STR profile, some loci may have results that give identical restricted and unrestricted LRs.
5.4.2.2.1 At a locus, a mixture with alleles P and Q is assumed to be from two contributors and displays no indications of allelic dropout. The peak height ratio is 50% (P allele taller). Across the entire profile, the mixture appears to be 2:1. The suspect in question is PP, and no other reference standards are being considered for inclusion.
The numerator of the LR calculation would assume the suspect’s contribution, meaning that the probability of observing results consistent with their genotype would be 1.0.
The second, unknown contributor must complete the mixture by having allele Q and nothing other than P or Q. If the assumed contributor (the suspect) is the minor contributor to the mixture, the possible second contributor genotypes included post-deconvolution might be PQ.
LR numerator = 2pq
Conversely, if the second contributor is the minor contributor, the possible second contributor genotypes included post-deconvolution might be QQ.
LR numerator = q2 + q(1-q)θ
The denominator of the LR calculation might assume that the mixture is a combination of two unknown contributors. The unknown contributors must have no alleles other than P or Q, and the combination of their genotypes must complete the detected mixture of P and Q. Based upon the relative peak height ratios and the overall mixture ratio, the restricted LR denominator might be limited to the following pairs of genotypes:
|
Major Contrib.
|
Minor. Contrib.
|
Combined Probability
|
|---|---|---|
|
PP
|
QQ
|
[p2 + p(1-p)θ] * [q2 + q(1-q)θ]
|
|
PQ
|
PP
|
2pq * [p2 + p(1-p)θ]
|
LR denominator = the sum of the possible combinations of genotypes (i.e., summing the two combined probabilities).
5.4.2.3 Additional formulae for restricted and unrestricted LRs can be found in Fung and Hu (2008).
Table 1 – Suitable Statistical Analyses for DNA Typing Results
The statistical methods listed in the table cannot be combined into one calculation. For example, combining RMP at one locus with a CPI calculation at a second locus is not appropriate. However, an RMP may be calculated for the major component of a mixture and a CPE/CPI for the entire mixture (as referred to in section 4.6.2).
|
Category of DNA Typing Result
|
RMP | CPE/CPI | LR (1) |
|---|---|---|---|
|
Single Source
|
•
|
•
|
|
|
Single Major Contributor to a Mixture
|
•
|
•
|
|
|
Multiple Major Contributors to a Mixture
|
• (2)
|
• (2)
|
•
|
|
Single Minor Contributor to a Mixture
|
•
|
• (3)
|
•
|
|
Multiple Minor Contributors to a Mixture
|
• (2)
|
• (3)
|
•
|
|
Indistinguishable Mixture
|
• (1)
|
•
|
•
|
(1) Restricted or unrestricted
(2) Restricted
(3) All potential alleles identified during interpretation are included in the statistical calculation
6. References and Literature Cited
Bär, W., Brinkmann, B., Lincoln, P., Mayr, W. R., and Rossi, U. (1994) DNA recommendations – 1994 report concerning further recommendations of the DNA Commission of the ISFH regarding PCR-based polymorphisms in STR (short tandem repeat) systems. Int. J. Legal Med. 107: 159-160.
Bär, W., Brinkmann, B., Budowle, B., Carracedo, A., Gill, P., Lincoln, P., Mayr, W. R., and Olaisen, B. (1997) DNA recommendations – further report of the DNA Commission of the ISFH regarding the use of short tandem repeat systems. Int. J. Legal Med. 110: 175-176.
Committee on DNA Forensic Science, National Research Council. An Update: The Evaluation of Forensic DNA Evidence. National Academy Press, Washington, DC, 1996.
DNA Advisory Board. Quality Assurance Standards for Forensic DNA Typing Laboratories, Forensic Sci. Comm. 2(3).
DNA Advisory Board (2000) Statistical and population genetic issues affecting the evaluation of the frequency of occurrence of DNA profiles calculated from pertinent population database(s). Forensic Sci. Comm. 2(3).
FBI Director (2009) Quality Assurance Standards for Forensic DNA Testing Laboratories.
Fung, W.K. and Hu, Y.-Q. (2008) Statistical DNA Forensics: Theory, Methods and Computation. Wiley: Hoboken, NJ.
Scientific Working Group on DNA Analysis Methods (SWGDAM). Short Tandem Repeat (STR) Interpretation Guidelines, Forensic Science Communications 2 (July 2000).
7. Additional Suggested Readings
Bill, M., Gill, P., Curran, J., Clayton, T., Pinchin, R., Healy, M., and Buckleton, J. (2005) PENDULUM-a guideline-based approach to the interpretation of STR mixtures. Forensic Sci. Int. 148: 181-189.
Buckleton, J.S., Evett, I.W., Weir, B.S. (1998) Setting bounds for the likelihood ratio when multiple hypotheses are postulated. Sci. Justice. 38: 23-26.
Buckleton, J.S., Curran, J.M., Gill, P. (2007) Towards understanding the effect of uncertainty in the number of contributors to DNA stains. Forensic Sci. Int. Genet. 1:20-28.
Buckleton, J.S. and Curran, J.M. (2008) A discussion of the merits of random man not excluded and likelihood ratios. Forensic Sci. Int. Genet. 2: 343-348.
Budowle, B., Chakraborty, R., Carmody, G., Monson, K.L. (2000) Source attribution of a forensic DNA profile. Forensic Sci. Commun. 2(3).
Budowle, B., Onorato, A.J., Callaghan, T.F., Della Manna, A., Gross, A.M., Guerrieri, R.A., Luttman, J.C., McClure, D.L. (2009) Mixture interpretation: defining the relevant features for guidelines for the assessment of mixed DNA profiles in forensic casework. J. Forensic Sci. 54: 810-821.
Clayton, T.M., Whitaker, J.P., Sparkes, R., Gill, P. (1998) Analysis and interpretation of mixed forensic stains using DNA STR profiling. Forensic Sci. Int. 91: 55-70.
Devlin, B. (1993) Forensic inference from genetic markers. Stat. Methods Med. Res. 2: 241-262.
Evett, I.W. Buffery, C., Willott, G., Stoney, D. (1991) A guide to interpreting single locus profiles of DNA mixtures in forensic cases, J. Forensic Sci. Soc. 31: 41–47.
Evett, I.W., Weir, B.S. (1998) Interpreting DNA Evidence. Sinauer: Sunderland, MA.
Gill, P., Sparkes, R., Kimpton, C. (1997) Development of guidelines to designate alleles using an STR multiplex system. Forensic Sci. Int. 89: 185-197.
Gill, P. (2002) Role of short tandem repeat DNA in forensic casework in the UK—past, present, and future perspectives. BioTechniques 32(2): 366-385.
Gill, P., Brenner, C.H., Buckleton, J.S., Carracedo, A., Krawczak, M., Mayr, W.R., Morling, N., Prinz, M., Schneider, P.M., Weir, B.S. (2006) DNA commission of the International Society of Forensic Genetics: Recommendations on the interpretation of mixtures. Forensic Sci. Int. 160: 90-101.
Gill, P., Brown, R.M., Fairley, M., Lee, L., Smyth, M., Simpson, N., Irwin, B., Dunlop, J., Greenhalgh, M., Way, K., Westacott, E.J., Ferguson, S.J., Ford, L.V., Clayton, T., Guiness, J. (2008) National recommendations of the technical UK DNA working group on mixture interpretation for the NDNAD and for court going purposes. FSI Genetics 2(1): 76–82.
Gill, P., Puch-Solis, R., Curran, J. (2009) The low-template DNA (stochastic) threshold-its determination relative to risk analysis for national DNA databases. Forensic Sci. Int. Genet. 3: 104-111.
Gill, P. and Buckleton, J. (2009) A universal strategy to interpret DNA profiles that does not require a definition of low-copy-number. Forensic Sci. Int. Genet. (in press).
Graham, E.A.M. and Rutty, G.N. (2008) Investigation into “normal” background DNA on adult necks: implications for DNA profiling of manual strangulation victims. J. Forensic Sci. 53: 1074-1082.
Ladd, C., Lee, H.C., Yang, N., Bieber, F.R. (2001) Interpretation of complex forensic DNA mixtures. Croatian Med. J. 42(3): 244-246.
Moretti, T.R., Baumstark, A.L., Defenbaugh, D.A., Keys, K.M., Budowle, B. (2001) Validation of short tandem repeats (STRs) for forensic usage: performance testing of fluorescent multiplex STR systems and analysis of authentic and simulated forensic samples. J. Forensic Sci. 46: 647-660.
Moretti, T.R., Baumstark, A.L., Defenbaugh, D.A., Keys, K.M., Budowle, B. (2001) Validation of STR typing by capillary electrophoresis. J. Forensic Sci. 46: 661-676.
Morling N., Bastisch, I., Gill, P., Schneider, P.M. (2007) Interpretation of DNA mixtures – European consensus on principles. Forensic Sci. Int. Genet. 1(3): 291–292.
Perlin, M. W. and Szabady, B. (2001) Linear mixture analysis: a mathematical approach to resolving mixed DNA samples. J. Forensic Sci. 46(6): 1372-1378.
Perlin, M.W., Kadane, J.B., Cotton, R.W. (2009) Match likelihood ratio for uncertain genotypes. Law, Probability and Risk 8(3):289-302.
Schneider, P.M., Gill, P., Carracedo, A. (2006) Editorial on the recommendations of the DNA commission of the ISFG on the interpretation of mixtures. Forensic Sci. Int. 160: 89.
Schneider, P.M., Fimmers, R., Keil, W., Molsberger, G., Patzelt, D., Pflug, W., Rothämel, T., Schmitter, H., Schneider, H., Brinkman, B. (2009) The German Stain Commission: recommendations for the interpretation of mixed stains. Int. J. Legal Med. 123: 1-5; originally published in German in 2006 Rechtsmedizin 16:401-404.
Stringer, P., Scheffer, J.W., Scott, P., Lee, J., Goetz, R., Ientile, V., Eckhoff, C., Turbett, G., Carroll, D., Harbison, S. (2009) Interpretation of DNA mixtures—Australian and New Zealand consensus on principles. Forensic Sci. Int. Genet. 3: 144-145.
Tomsey, C.S., Kurtz, M., Flowers, B., Fumea, J., Giles, B., Kucherer, S. (2001) Case work guidelines and interpretation of short tandem repeat complex mixture analysis. Croatian Med. J. 42(3): 276-280.
Torres, Y., Flores, I., Prieto, V., Lopez-Soto, M., Farfan, M.J., Carracedo, A., Sanz, P. (2003) DNA mixtures in forensic casework: a 4-year retrospective study. Forensic Sci. Int. 134: 180-186.
Tvedebrink, T., Eriksen, P.S., Mogensen, H.S., Morling, N. (2009) Estimating the probability of allelic drop-out of STR alleles in forensic genetics. FSI Genetics 3: 222-226.
Van Nieuwerburgh, F., Goetghebeur, E., Vandewoestyne, M., Deforce, D. (2009) Impact of allelic dropout on evidential value of forensic DNA profiles using RMNE. Bioinformatics 25: 225-229.
Wang, T., Xue, N., Birdwell, J.D. (2006) Least-squares deconvolution: a framework for interpreting short tandem repeat mixtures. J. Forensic Sci. 51(6): 1284-1297.
Weir, B.S., Triggs, C.M., Starling, L., Stowell, L.I., Walsh, K.A.J., Buckleton, J.S. (1997) Interpreting DNA mixtures. J. Forensic Sci. 42(2): 213-222.
Whitaker, J.P., Cotton, E.A., Gill, P. (2001) A comparison of the characteristics of profiles produced with the AmpFlSTR SGM Plus multiplex system for both standard and low copy number (LCN) STR DNA analysis. Forensic Sci. Int. 123: 215-223.
Wickenheiser, R.A. (2006) General guidelines for categorization and interpretation of mixed STR DNA profiles. Canadian Society of Forensic Science Journal 39(4): 179-216.
Glossary for this document
Allelic dropout: failure to detect an allele within a sample or failure to amplify an allele during PCR.
Analytical threshold: the minimum height requirement at and above which detected peaks can be reliably distinguished from background noise; peaks above this threshold are generally not considered noise and are either artifacts or true alleles.
Artifact: a non-allelic product of the amplification process (e.g., stutter, non-templated nucleotide addition, or other non-specific product), an anomaly of the detection process (e.g., pull-up or spike), or a by-product of primer synthesis (e.g., “dye blob”).
Coincidental match: a match which occurs by chance.
Composite profile: a DNA profile generated by combining typing results from different loci obtained from multiple injections of the same amplified sample and/or multiple amplifications of the same DNA extract. When separate extracts from different locations on a given evidentiary item are combined prior to amplification, the resultant DNA profile is not considered a composite profile.
Conditional: an interpretation category that incorporates assumption(s) as to the number of contributors.
CPE: combined probability of exclusion; produced by multiplying the probabilities of inclusion from each locus and subtracting the product from 1 (i.e., 1-CPI).
CPI: combined probability of inclusion; produced by multiplying the probabilities of inclusion from each locus (i.e., 1-CPE).
Deconvolution: separation of contributors to a mixed DNA profile based on quantitative peak height information and any underlying assumptions.
Deduced: inference of an unknown contributor’s DNA profile after taking into consideration the contribution of a known/assumed contributor’s DNA profile.
Differential degradation: a DNA typing result in which contributors to a DNA mixture are subject to different levels of degradation (e.g., due to time of deposition), thereby impacting the mixture ratios across the entire profile.
Distinguishable mixture: a DNA mixture in which relative peak height ratios allow deconvolution of the profiles of major/minor contributor(s).
Evidence sample: also known as “questioned sample.”
Exclusion: a conclusion that eliminates an individual as a potential contributor of DNA obtained from an evidentiary item based on the comparison of known and questioned DNA profiles (or multiple questioned DNA profiles to each other).
Guidelines: a set of general principles used to provide directions and parameters for decision making.
Heterozygote: an individual having different alleles at a particular locus; usually manifested as two distinct peaks for a locus in an electropherogram.
Homozygote: an individual having the same (or indistinguishable) alleles at a particular locus; manifested as a single peak for a locus in an electropherogram.
Inclusion: a conclusion for which an individual cannot be excluded as a potential contributor of DNA obtained from an evidentiary item based on the comparison of known and questioned DNA profiles (or multiple questioned DNA profiles to each other).
Inconclusive/uninterpretable: an interpretation or conclusion in which the DNA typing results are insufficient, as defined by the laboratory, for comparison purposes.
Indistinguishable mixture: a DNA mixture in which relative peak height ratios are insufficient to attribute alleles to individual contributor(s).
Intimate sample: a biological sample from an evidence item that is obtained directly from an individual’s body; it is not unexpected to detect that individual’s allele(s) in the DNA typing results.
Known sample: biological material for which the identity of the donor is established and used for comparison purposes (referred to as a “K”).
Likelihood ratio (LR): the ratio of two probabilities of the same event under different hypotheses; typically the numerator contains the prosecution’s hypothesis and the denominator the defense’s hypothesis.
Major contributor(s): an individual(s) who can account for the greater portion of the DNA in a mixed profile.
Masked allele: an allele of the minor contributor that may not be readily distinguishable from the alleles of the major contributor or an artifact.
Minor contributor(s): an individual(s) who can account for the lesser portion of the DNA in a mixed profile.
Mixture: a DNA typing result originating from two or more individuals.
Mixture ratio: the relative ratio of the DNA contributions of multiple individuals to a mixed DNA typing result, as determined by the use of quantitative peak height information; may also be expressed as a percentage.
Noise: background signal detected by a data collection instrument.
No results: no allelic peaks detected above the analytical threshold.
Obligate allele: an allele in a mixed DNA typing result that is (a) foreign to an assumed contributor, or (b) based on quantitative peak height information, determined to be shared with the assumed contributor.
Partial profile: a DNA profile for which typing results are not obtained at all tested loci due, for example, to DNA degradation, inhibition of amplification, and/or low-quantity template.
Peak height ratio (PHR): the relative ratio of two alleles at a given locus, as determined by dividing the peak height of an allele with a lower relative fluorescence unit (RFU) value by the peak height of an allele with a higher RFU value, and then multiplying this value by 100 to express the PHR as a percentage; used as an indication of which alleles may be heterozygous pairs and also in mixture deconvolution.
Probability of exclusion (PE): the percentage of the population that can be excluded as potential contributors to a DNA mixture.
Probability of inclusion (PI): the percentage of the population that can be included as potential contributors to a DNA mixture; also known as “Random Man Not Excluded.”
Questioned sample: biological sample recovered from a crime scene or collected from persons or objects associated with a crime (referred to as a “Q”).
Random Match Probability (RMP): the probability of randomly selecting an unrelated individual from the population who could be a potential contributor to an evidentiary profile.
Reference sample: also known as “known sample.”
Restricted: referring to a statistical approach conditioned on the number of contributors and with consideration of quantitative peak height information and inference of contributor mixture ratios; used to limit the genotypic combinations of possible contributors.
Signal-to-noise ratio: an assessment used to establish an analytical threshold to distinguish allelic peaks (signal) from background/instrumental noise.
Single-source profile: DNA typing results determined to originate from one individual based on peak height ratio assessments and the number of alleles at given loci.
Source attribution: a declaration which identifies an individual as the source of an evidentiary profile to a reasonable degree of scientific certainty based on a single-source or major contributor profile.
Stochastic effects: the observation of intra-locus peak imbalance and/or allele drop-out resulting from random, disproportionate amplification of alleles in low-quantity template samples.
Stochastic threshold: the peak height value above which it is reasonable to assume that, at a given locus, allelic dropout of a sister allele has not occurred.
Stutter: a minor peak typically observed one repeat unit smaller than a primary STR allele resulting from strand slippage during amplification.
Unrestricted: referring to a statistical approach performed without consideration of quantitative peak height information and inference of contributor mixture ratios; for CPE/CPI, this may or may not be conditioned on the number of contributors.
08.16.10